Biomedical Data
All data: has primary purpose (why it was collected) and may have secondary use (e.g. research in EMR data). It is important to clarify what the primary purpose is for secondary uses.
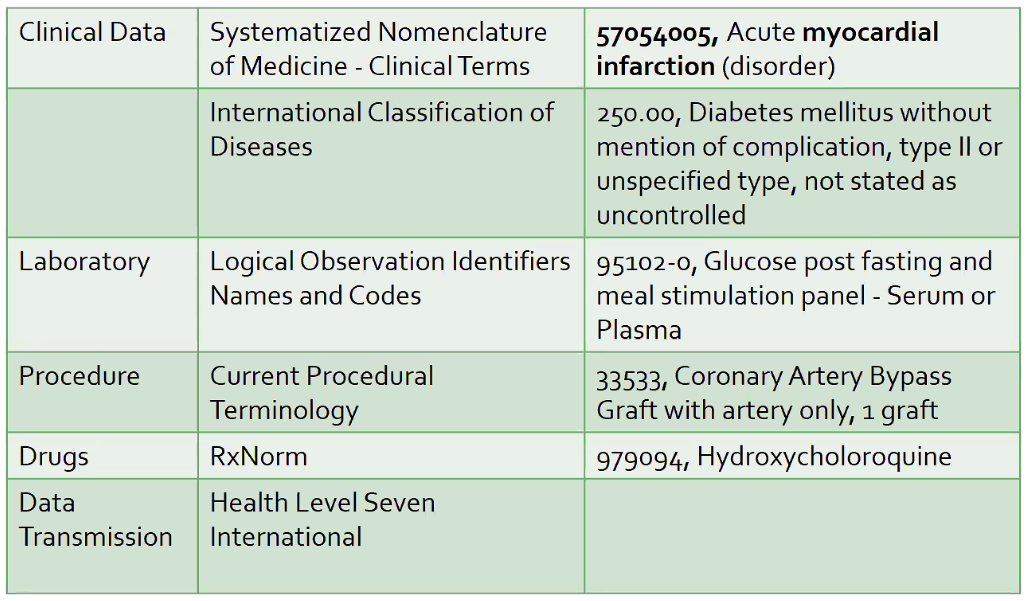
Types - Narrative (e.g. text), discrete (numeric codes – easiest to store, transmit, query, and compute), signals (ECG, ventilator data), Images (x-rays, hand drawn images), genomic/genetic (semistructured), exposures
Text mining = taking instructured text and creating discrete data. An example is natural language processing.
Structured data, all have 4 attributes: who, what, which, when
Computability – depends on heterogeneity (many types of data; lack of standard canonical form, meaning a preferred representation that supersedes other aliases), Volume, poor mathematical characterization
What makes big data: Volume (size, amont), velocity (rate at which data comes in, needs used), variety, veracity (what is the quality, truthfulness), value
Information is data with meaning. Knowledge is information that is justifiably believed to be true. Note: data is increasing quickly, not clear that knowledge is
Giving structure to data: Standards / metadata / ontologies
Hierarchies: flat – terminology; very tiered – ontology.
Example Terminologies
NIH – very interested in capturing information about studies (e.g. clinicaltrials.gov) = meta-data about the study itself.
PHI
Regulations
Regulatory Agencies: OHRP, FDA. Important Documents: Belmont Report, Code of Federal Regulations (CFR), International Conference on Harmonization (ICH), Good Clinical Practice (GCP)
Human subjects => use 45 CFR part 46 (protection of human subjects) to determine what regulations apply
Data with patient identifiers = the following things.
4 ways to describe the level of patient identification
- Anonymous data - patient ID was removed at the time of collection
- Anonymized data - initially has identifiers, which are subsequently irrevocably removed
- De-identified data - collected with identifiers that are subsequently encoded or encrypted. Could be re-identified if stipulated (e.g. by IRB)
- Identified data - patient identified; generally requires consent.
De-identified data is considered 'minimal risk' data - only risk of data breach is loss of confidentiality. Still may be exempt via IRB.
Data where identifiers are needed, but then subsequently destroyed, do require a waiver of informed consent (rather than possible exemption), or data-use agremements (e.g. in the case where it's possible to re-identify, or some PHI is needed)
EMR data
Primary use: patient care. Secondary use: research.
Missingness is ubiquitous when data not collected as part of research protocols.
Tension in how data is recorded - Expressiveness is helpful in clinical documentation (primary use), whereas precision (limiting choices) is helpful in research and billing (secondary uses)
Clinical Data Repositories and Warehouses
Data repository = contains all of the raw information - usually transactional and optimized to retrieve or enter single patient updates; Warehouse implies some processing to allow quicker access (e.g. searching, queries, etc.) across patient populations.
This can be done with 1.) local storage but minimal processing (warehouse not as optimized), 2.) significant reprocessing into warehouse storage tables to allow optimal queries, or 3.) distributed - with data remaining in the repository (e.g. EMR), meaning slow query performance.
Data mart: subset of warehouse that is presented to investigators -> some segment of the warehouse databases is exported to be more consistent (not constantly updating) and readable/improved interface
Data flow -> usually 'data dumps' from the transactional system occur and update (or more often, totally overwrite) the warehouse dataset.
Warehouses usually use the Entity-attribute-value schema:
- Observations (defined observer, patient, event, modifiers) stored in a Fact table.
- Patient tables record patients in the database and their common attributes.
- Event tables - can be defined at different levels, e.g. doctors visit, lab collection event, etc.
- Observer tables - generally providers (though could be machines)
- Concept table used to link facts to human readable descriptions.
Metadata: ICD (dx), CPT (procedure), NDC (national drug codes), LOINC (tests)
Historically, administrative/billing databases and research data warehouses (often de-identified, slower refresh cycles, integration with other data sources and types of info like images) were separate.
Clinical Trial Management Systems
Goal: support management of all aspects of a clinical trial
- Traditional from commercial vendors: Oracle Clinical, Phase Forward Clinitrials
- Web-based solutions
- Add-ons to Office, Cognos
- Open access software: TrialDb, OpenClinica?
Attempt to balance consistency+compliance w customizability
Artificial Intelligence / Machine Learning
Applications
- Reasoning (aka inferring facts from knowledge and data)
- Problem-solving: reaching a goal through a sequence of steps/actions
- Knowledge representation: designing representation that capture information about the world that can be used to solve complex-problems
- Planning: to set goals and achieve them
- Learning: computer algorithms that improve with experience
- Natural language processing: ability to read, understand, and generate human language
In general, sensitivity is much easier to achieve than specificity in classification tasks
Machine Learning
Supervised vs unsupervised learning
Supervised learning
assumes that you know a ground truth examples that the machine can learn based on (termed labeled data) -> then this knowledge is applied on unknown samples.
One challenge - the features that the algorithm will use to classify patients are an important choice and should represent the pertinent to the classification problem.
Output can be discrete (classification task) or continuous (regression task).
Unsupervised learning
Does not have labeled outputs - no reference standards. Goal is to infer the natural structure within a set of data points. Tries to classify objects based on similarity.
Often helpful in generating hypotheses that are not intuitively obvious.
Clustering (by K-means) is the workhorse algorithm.
Data Management
Technology + Research/protocols and process (aka workflows)
- initially collected documents = source documents, must conform to study design ie. IRB. Auditors should be able to reconstruct a participants course
Data domain: abstract representation of knowledge or activity - elements when aggregated contribute to understanding/knowledge. E.g. demographic data, eligibility criteria, treatments, PROs, follow-up etc.
Data Editor: validates, creates, or edits data Data Steward: holds the data - responsible. Generally the PI Data Owner = approves data before publication. Data consumer = statistician, NIH, etc.
Tasks:
- Data capture: need to make sure that you are getting plausible values, excluding errors. Need to support clean data without interfering with clinical work flows.
- Data harmonization: translating code and metadata such that data from multiple sources that refer to the same thing are consolidated (requires a mapping between different datasources) - e.g. BCx; Blood Culture; Culture, Blood; 747233; 5862 all refer to same thing
- Data modeling: how is the data organized so that it fits the purpose of the research study
'Integration' = the process going from raw data -> useful information.
- Federation = data collected from distinct databases without copying/transferring the data itself
- Aggregation = compiling data from different datasets into a new, combined dataset for processing and analysis
Databases
Generally interfaced through a Database management system, which works with several applications (or directly)
Vs. a spreadsheet - enforces rules about the structure of the data; adds functions that protect the data.
Vs. a filesystem - data redundancy and consistency; ease of accessing data; easier to add rules to enforce data integrity; multiple users (performance, avoid multiple read/write problems); security.
Redcap
Application that sits on top of a database to facilitate data collection
Can be PHI (e.g. HIPAA and other statutes) compliant
Uses:
- surveys (participant doesn't log in, just enters data), forms (authorized user enters data)
- databases
- allows validation of entries/data quality assurance; branching/skip logic; uploads
- can export to other packages / formats by running 'reports' - there are some limited statistical analysis. This can be done in a de-identified format.
- can embed consent
Field types: data/time, short strings, radio buttons or drop downs, checkbox, matrix, visual analog, yes/no => QA
Data dictionary - can use an online designer to create questions for data collection
Redcap can be a good place to store small dataset. (On Ubox can be a good way also)
Protected Environment at CHPC
Built for storing larger databases in a HIPAA required way - requires authentication and log in to virtual machine. Data stays there and computation occurs within the environment.
https://www.chpc.utah.edu/resources/ProtectedEnvironment.php
##Data sources
Secondary:
- single institution (e.g. via EDW, [email protected])
- research network
Utah Population Database Utah Department of Health (all claims payor database, public health indicator based information system aka IBIS)
TriNetX
Federated architecture, but harmonized into a shared data-set. Mostly EHR data, but adding other information.
Use cases:
- get de-identified data for preliminary data e.g. to support grants
- initiate trials across institutions if you find you need more people
ACT network
CTSA Federated Network as part of NCATS funding, which has now finished. So, it is stagnant, TriNetX supplanted.
PCORI
Patient centered outcome research network - Great Plains collaborative. Rachel Hess is in charge. No query interface.
Data Extraction
Adding domain specific logic and concepts to create subsets of data for research use.
- e.g. eligibility criteria (inclusion/exclusion): the rules (logic, including temporality) that select for a set of individuals is called a phenotype.
Data Modeling
Take extracted data and put in a place where it can be analyzed and computed.
OMOP (observational medical outcomes partnership) common data model = most common
[ TriNetX is essentially in OMOP ]
[ ] The Book of Ohdsi
Data Integration
Many platforms available to help bring together (if multiple sources).
Federation: data stays in separate database but requests are harmonized Aggregation: data is combined into a large database, then analyzed
Process Modeling
Study management: e.g. open clinica (open source), RED Cap